The work in my group mostly focuses on membrane proteins and peptides targeting them with a touch of small molecule – protein interactions. We combine traditional computational chemistry and structural biology tools such as Rosetta and molecular dynamics simulations with state-of-the-art machine-learning-based methods to develop methods or applications that can be used for the investigation of ligand and protein structure and function. Below is a list of the available projects and the required skills to work on these projects:
Development of a framework for the prediction of membrane protein – membrane interactions with Rosetta
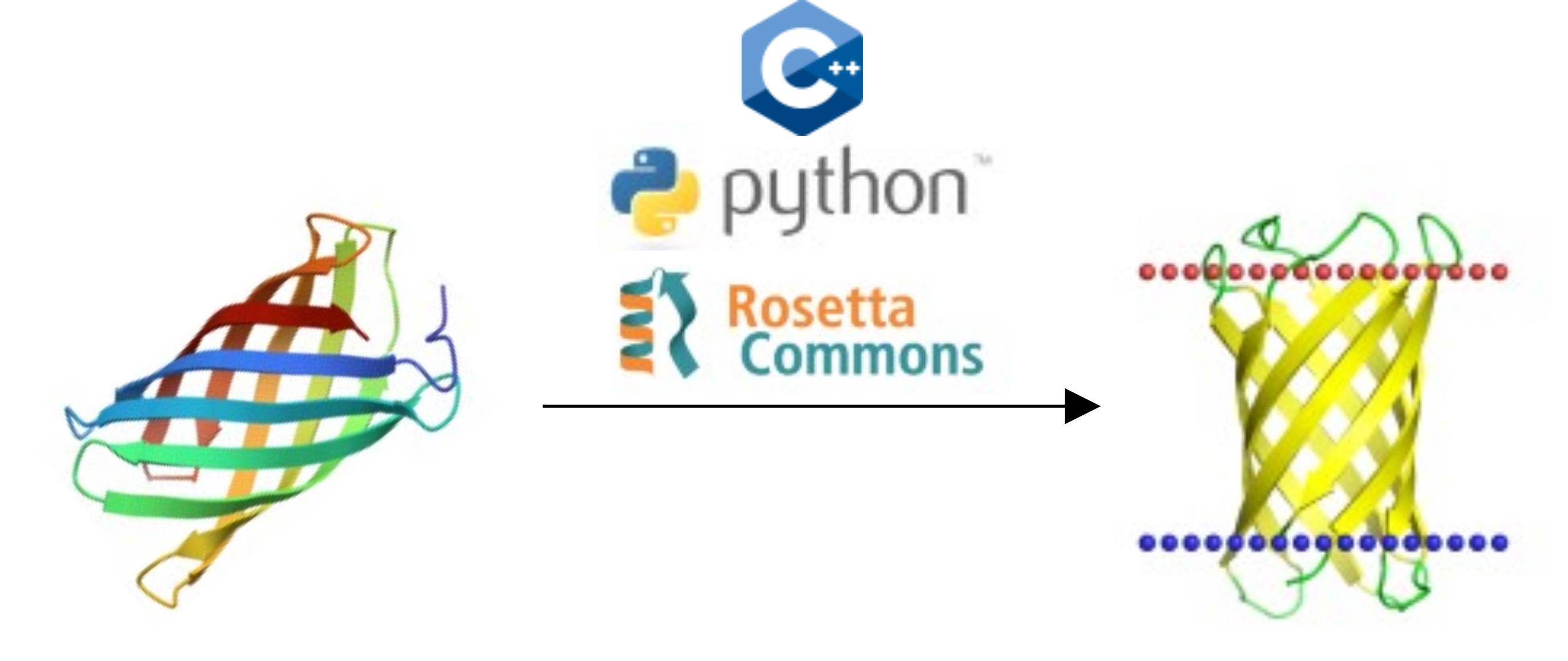
In this project, our aim is to develop and benchmark a protocol for accurate placement of membrane proteins in membranes. Placement of proteins in membrane models is a prerequisite of many calculations involving membrane proteins and the resulting framework will be frequently used by the researchers working in this field. You will have the chance to put your programming skills (Python or C++) into action in order to create and test simple algorithms to sample the energy landscape of proteins in and on membranes. This project presents an opportunity for students with programming skills to learn more about protein structure and function, frequently used structure modeling tools like Rosetta, and scientific method development.
Required Skills
- An interest in applying programming to scientific questions.
- Motivation to learn about protein structure prediction and manipulation tools.
- Basic understanding of Bash and Python or C++ (variables, for and if loops, functions, classes).
Investigation of small molecule – nicotinic acetylcholine receptor (nAChR) investigations from a computational perspective
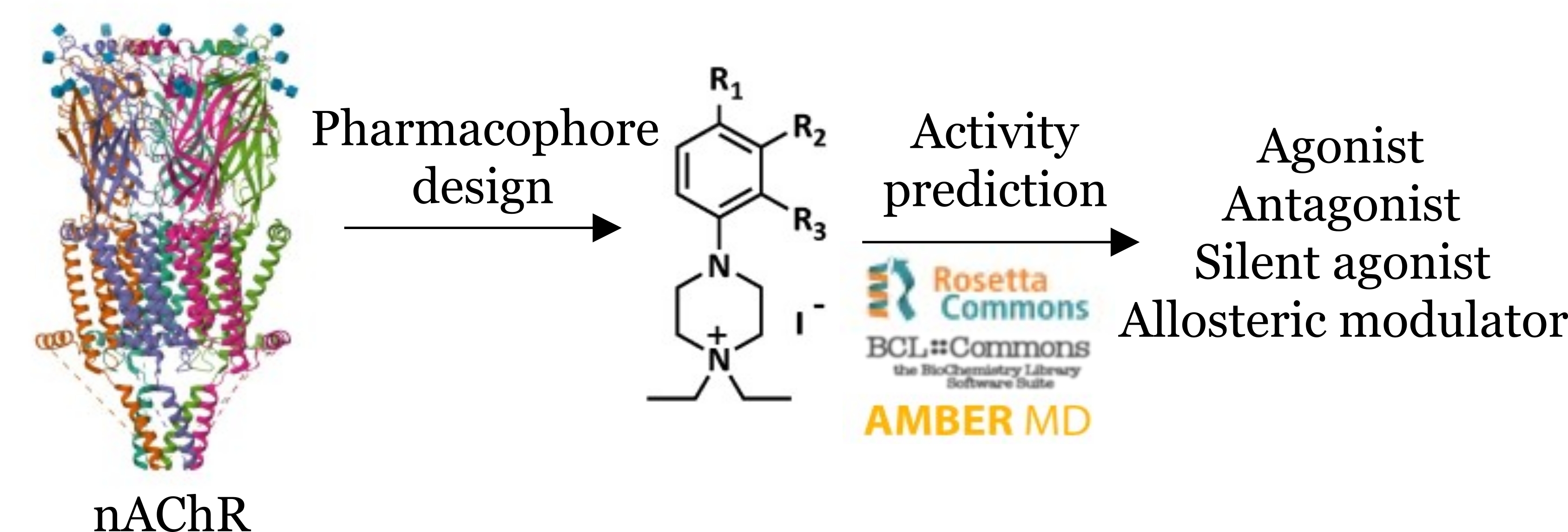
This project focuses on understanding how and why similar drugs differentially target different proteins belonging to the same family. You will have the opportunity to learn drug discovery tools while exploring how drug molecules work at a family of receptors important for many neurological and immune diseases. Working on this project will help you learn the basics of doing chemistry with computers and technical skills involving basic programming with Bash and Python. You will also learn more about drug – protein interactions and how ion-channels function while you put your theoretical knowledge on the principles of medicinal chemistry to work.
Required Skills
- An interest in computational drug development tools.
- Motivation to learn about ligand gated ion-channel pharmacology.
- Basic knowledge of organic chemistry and biochemistry (functional groups and their effects, protein structure basics, how inter- and intra-molecular interactions work).
Development and application of computational tools for the modeling of peptide –protein interactions through machine-learning
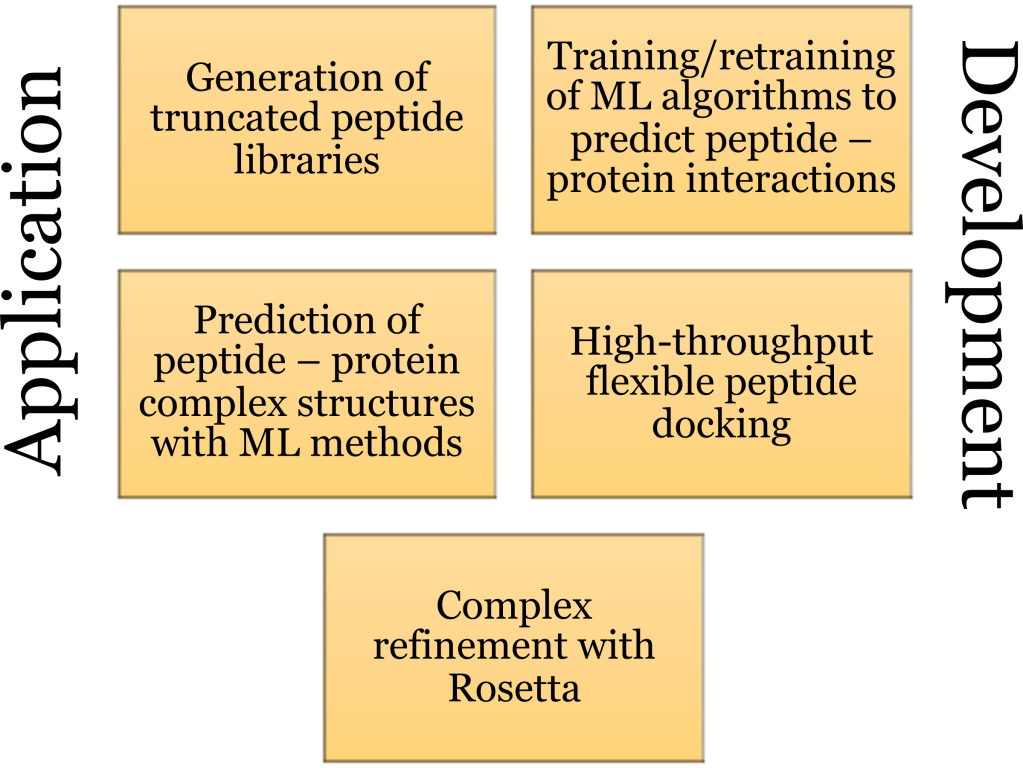
Peptides pose exciting targets as drugs for the treatment of many diseases with high selectivity. Accurate modeling of peptide – protein interactions is key to the development of peptides with promising therapeutic properties. In this project, you will have the opportunity to learn about the use of machine learning and other protein/peptide modeling tools in computational chemistry from different perspectives. Part of this project focuses on application of existing methodology for less experienced students and another part focuses on cutting-edge method development for more experienced students. The ultimate goal of the project is the development of peptide binders targeting membrane proteins de novo (i.e., not existing in nature) or using naturally occurring peptide scaffolds.
Required Skills
- An interest in method and/or protocol development using protein structure prediction tools.
- Basic (application part) or advanced (development part) understanding of machine learning algorithms and their applications.
- Basic (application part) or advanced (development part) understanding of programming with Python.